rbasset <- function(n){
Z <- rbinom(n, size=1, prob=0.6) # Z=1 <- Female
ifelse(Z, rnorm(n, mean=55, sd=sqrt(10)),
rnorm(n, mean=65, sd=sqrt(10)))
}
rbasset(1)[1] 53.06954Michael Weylandt
January 23, 2026
\[\newcommand{\bX}{\mathbf{X}}\newcommand{\bx}{\mathbf{x}}\newcommand{\R}{\mathbb{R}}\newcommand{\P}{\mathbb{P}}\]
This week, we begin our study of classifiers with an analysis of generative classifiers. While many generative classifiers have appeared in the literature, they can essentially all be derived from a standard formula. We will discuss this standard formula first and then turn our attention to specific named instances.
Recall from STA 9715 that a (discrete) mixture model is a distribution of the form:
\[X \sim \begin{cases} X_1 & Z = 1 \\ X_2 & Z = 2 \\ \dots \\ X_K & Z=K \end{cases} \text{ where } \textsf{supp}(Z) = \{1,\dots, K\} \text{ and } X_i \sim \mathcal{P}_i \text{ for all $i$ and } Z \perp X_i, X_i \perp X_j\, (i\neq j)\]
That’s quite a bit to unpack, so let’s take it piece by piece: the random variable \(X\) has a \(K\)-component mixture distribution if we can sample \(X\) by generating \(K\) independent random variables \(\{X_1, \dots, X_K\}\) (with different distributions) and then selecting one of them at random.
Let’s look at a concrete version of this: suppose we have a population of male and female basset hounds that is 60% female. The weight of male basset hounds follows a \(\mathcal{N}(65, 10)\) distribution while females have weight distributed as \(\mathcal{N}(55, 10)\). The weight of a randomly selected basset can be sampled as
rbasset <- function(n){
Z <- rbinom(n, size=1, prob=0.6) # Z=1 <- Female
ifelse(Z, rnorm(n, mean=55, sd=sqrt(10)),
rnorm(n, mean=65, sd=sqrt(10)))
}
rbasset(1)[1] 53.06954If we collect the weights of many basset hounds, we see that the resulting distribution is not normally distributed:
hist(rbasset(10000), breaks=50, main="Distribution of Basset Hound Weights")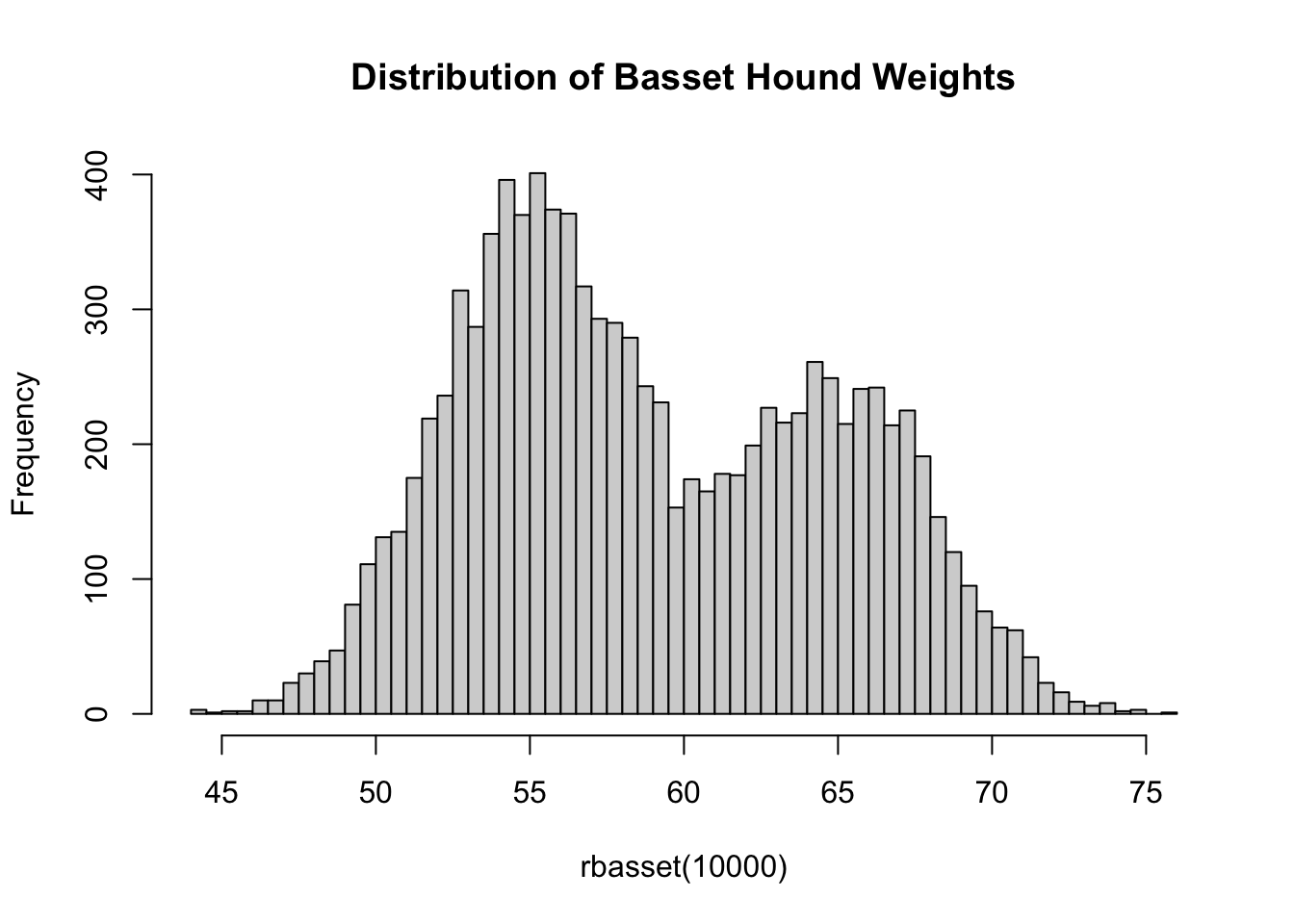
In fact, this is a bimodal distribution with each ‘lump’ representing males or females.1 Distributions of this form can often be written more compactly using a ‘convex combination’ notation:
\[\text{Basset} \sim \frac{2}{5}\mathcal{N}(65, 10) + \frac{3}{5}\mathcal{N}(55, 10)\]
The above notation can be interpreted as specifying a draw from the first distribution with probability \(2/5 = 40\%\) and from the second with probability \(3/5 = 60\%\); that is, it’s a particular 2-component mixture of normals.
In specifying this mixture, the probabilities of each component must be non-negative and must sum to 1; these are exactly the same restrictions we put on a convex combination of points earlier in the course. In fact, the set of mixture distributions can be consider the ‘convex set’ containing all of its individual components.
Now suppose you are on a walk and you meet a very friendly 62 pound basset hound. You of course want to pet this marvelous dog, but you need to ask the owner’s permission first: in doing so, should you ask “may I pet him?” or “may I pet her?”
While the basset hound won’t care if you misgender, you recall that you can use Bayes’ Rule to ‘invert’ the mixture distribution. That is, while the mixture distribution normally goes from sex to weight, Bayes’ rule will let us go from weight back to sex.
Specifically, in this case, Bayes’ rule tells us:
\[\P(\text{Female}|\text{Weight=62}) = \frac{\P(\text{Weight=62}|\text{Female})\P(\text{Female})}{\P(\text{Weight=62}|\text{Female})\P(\text{Female}+\P(\text{Weight=62}|\text{Male})\P(\text{Male}))}\]
The math here is a bit nasty numerically, but we can do it in R quite simply:
p_female <- 0.6
p_male <- 1-p_female
p_62_female <- dnorm(62, mean=55, sd=sqrt(10))
p_62_male <- dnorm(62, mean=65, sd=sqrt(10))
p_female_62 <- (p_62_female * p_female) / (p_62_female * p_female +
p_62_male * p_male)
paste0("There is a ", round(p_female_62 * 100, 2), "% chance that basset is a female.")[1] "There is a 16.87% chance that basset is a female."Given that calculation, you take a chance and ask whether you can pet him.
What have we done here? We have built a basset-sex classifier from a combination of the population distribution and Bayes’ rule. This is the essence of generative classifiers - if we know the distribution, accurate probabilistic classification is a straightforward application of Bayes’ rule. In practice, we don’t know the distribution a priori and it must be estimated from data. Different generative classifiers essentially are just different ways of estimating the population (mixture) distribution.
Let’s formalize our approach for a two-class classifier though the extension to multi-class is simple enough. Given a training set
\[\mathcal{D}_{\text{Train}} = \{(\bx_1, y_1), (\bx_2, y_2), \dots, (\bx_n, y_n)\}\]
we assume \(\bX\) follows a two-component mixture where \(Y\) is the mixing variable:
\[\bX | Y=0 \sim \mathcal{P}_0 \text{ and } \bX | Y=1 \sim \mathcal{P}_1\]
Clearly to fully specify this distribution, we need three things:
The distribution of \(Y\) is relatively straightforward: since it only takes values \(\{0, 1\}\) it must have a Bernoulli distribution and hence the only thing we need to estimate is the probability, \(\pi_1\), that \(Y=1\). The simplest approach to estimating the Bernoulli probability is to simply take the empirical probability from the training data set, i.e., the fraction of observations where \(Y=1\), but this can also be estimated using knowledge of a broader data set or a different population than the training data. For instance, if a data set is known to overindex on \(Y=1\), e.g., because it contains patients who self-reported some disease, it may be more appropriate to estimate \(p\) based on the population-wide prevalance of the disease. These concerns are typically rather problem specific.
Estimation of \(\mathcal{P}_0, \mathcal{P}_1\) may be more difficult. If the training data is extremely large, we may use a non-parametric estimator, e.g., a multivariate histogram, but in general we will need to use some sort of restrictive (parametric) model. The most commonly used generative classifiers assume some sort of multivariate normal structure, but you can create ‘custom’ classifiers appropriate to specific domains.
For now, let us assume \(\mathcal{P}_{0}, \mathcal{P}_1\) have been estimated and that they have PDFs \(p_0(\cdot), p_1(\cdot)\). Applying Bayes’ rule as before, with prior (baseline) probabilities \(\pi_0, \pi_1 = 1-\pi_0\), we get the following probabilities
\[\begin{align*} \P(Y = 1 | \bx) &= \frac{\P(\bx | Y = 1)\P(Y=1)}{\P(\bx | Y = 0)\P(Y=0)+\P(\bx | Y = 1)\P(Y=1)} \\ &= \frac{p_1(\bx)\pi_1}{p_0(\bx)\pi_0 + p_1(\bx)\pi_1} \end{align*}\]
If we are happy with a probabilistic classifier, we are essentially done. If we want a true binary classifier, i.e., a \(\{0, 1\}\) valued ‘point predictor’, we also need to set a classification threshold, \(\overline{p}\), to obtain:
\[\delta(\bx) = \begin{cases} 0 & \frac{p_1(\bx)\pi_1}{p_0(\bx)\pi_0 + p_1(\bx)\pi_1} < \overline{p} \\ 1 & \text{ otherwise} \end{cases}\]
\(\overline{p}\) can be chosen to optimize a relevant performance metric (if you practice good sample splitting hygiene) or via a full decision theoretic analysis, as discussed last week.
That’s basically it!
As mentioned above, the ‘popular’ generative classifiers are essentially defined by specific multivariate normality assumptions on \(\mathcal{P}_0, \mathcal{P}_1\). Let us review these in ascending order of complexity.
Recall that a multivariate normal distribution is uniquely identified by its mean and (co)variance (matrix), just like a standard univariate normal distribution. When estimating means, we will simply use the class means for each classifier, though James-Stein time estimators could be used for a (likely imperceptible) improvement. Estimation of variances is a harder task: to estimate the full covariance matrix of \(p\) random variables (or a random \(p\)-vector), we must estimate:
Making this even harder, we have to deal with the fact that the “variance of the variance”, i.e., the kurtosis, is typically quite large. While we do not focus on high-dimensional covariance estimation in these notes, it is quite a rich subject: if you want dig further into it, the Ledoit-Wolf estimator is a good place to start.
In Naive Bayes, we assume \(\mathcal{P}_0, \mathcal{P}_1\) are multivariate normal distributions with no correlation among the features and a common covariance across classes.2 This significantly simplifies the covariance estimation problem, since we now have all \(n\) samples available to estimate only \(p\) variances. Our estimates are given as follows:
\[\hat{\mu}_0 = \frac{1}{|\mathcal{C}_0|} \sum_{i \in \mathcal{C}_0} \bx_i\]
where \(\mathcal{C}_0\) is the set of observations from class 0 and
\[\hat{\mu}_1 = \frac{1}{|\mathcal{C}_1|} \sum_{i \in \mathcal{C}_1} \bx_i.\] The variance estimates are a bit trickier:
\[\hat{\Sigma}_{ii} = \frac{1}{n}\left(\sum_{j\in\mathcal{C}_0} (x_{ji} - \hat{\mu}_{0i})^2 + \sum_{j\in\mathcal{C}_1} (x_{ji} - \hat{\mu}_{1i})^2\right)\] This looks a bit nasty, but it’s not too hard: we take the standard ‘average squared difference’ estimator of the variance, but instead of using the overall mean, we subtract the estimated mean from the relevant class. Note that here we are only estimating \(\Sigma_{ii}\), the diagonal elements, and not a full matrix \(\Sigma\).
Naive Bayes classifiers are particularly important to the history of ML as they were used for one of the first and most influential spam filters.
Generalizing a bit, we can relax the assumption that the covariance matrix \(\Sigma\) is diagonal and only assume that it is constant across the two classes. Here,
Suppose we have
Note that a mixture of normals is not always multimodal (e.g., if the two means are very close together), but a mixture of normals is a common model for multimodal data.↩︎
Sometimes, we go further and assume the standard deviations of each feature are the same: this assumption is typically not made explicitly as such but is done ‘off-camera’ by pre-standardizing features.)↩︎